Water security is one of the leading issues in the world today. One way to detect contamination in water is by detecting E. coli. I built a monochromatic photometer to detect E. coli in water, and won bronze in the Canada-Wide Science Fair.
When water is contaminated with feces, there is a presence of E. coli in the water. While not all strains of E. coli are dangerous, the presence itself shows the water is not potable, which is why E. coli is universally tested for its presence in water. However, testing for E. coli and its concentration in the field is not an easy task. Spectrophotometry allows us to get an idea for concentrations of various contaminants in water, so is it possible to build one for field use?
I was to build a spectrophotometer. At the end of the day, this system is simply a light which shines through a sample, and a detector which detects the amount of light. Most spectrophotometers operate across a large spectrum, but they are also in the tens of thousands of dollars. Building a small spectrophotometer which only operates in a wavelength tailored to E. coli could save cost and make the device portable.
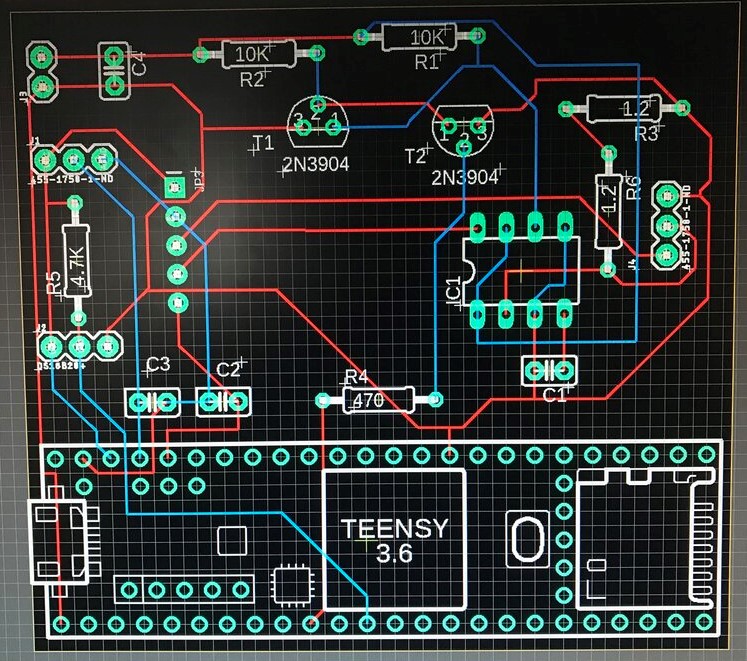
After further research, it seemed that E. coli absorbed high levels of light at UVC frequencies, and an LED of 260nm was selected. After this was selected, a light detection chip was selected to work in this UV frequency band. The control circuit design is below.
From here, putting together the circuit was a fairly simple task of soldering the components to the protoboard. I also printed an enclosure for the electronics. If you notice, there is a metal casing around the LED. This was for two reasons. First having this directed the light which is good for safety when dealing with high power UV LEDs. Second, it served as a heat sink, as I was not sure if the LED would be able to dissipate the generated heat by itself. In retrospect this likely was not needed due to the short operate period.
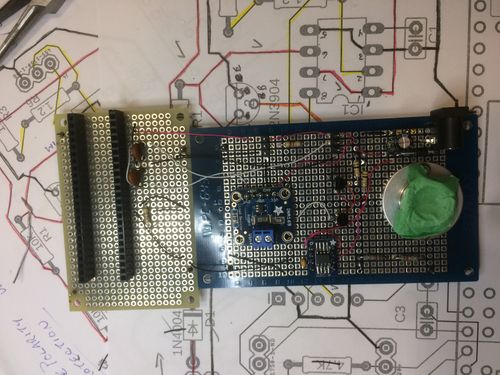

The final design ended up looking something like below (this is an older version), where the top could come off, and a water sample could be placed inside.

To test the spectrophotometer I built, I needed E. coli samples. I ended up connecting with a lab in Thunder Bay (where I am originally from), explained my hypothesis, and they allowed me to learn the E. coli culturing and testing process. The steps in this process were long, an excerpt from my submission to the Canada-Wide Science Fair is as follows:
"To make the bacteria cultures, strain K12 E. coli was ordered from Sigma-Aldrich. Nutrient broths were made using Lysogeny broth (LB) with 5 g of nutrient agar. A dried pellet from Sigma-Aldrich is added to 5 ml of water and is vortexed for one minute intervals for 5minutes. Then from those, 1 µL was added to each LB broth purity plate. From that a streak plate is made and grown for 24 - 48 hours at 37O C in an incubator..."
This goes on for a while, but I learned the process of working in a lab environment to work with Biosafety level 1 pathogens. Below is a picture from the E. coli plating process.

As a funny note, to me, E. coli when plated smells exactly like chicken noodle soup.
After the process of plating, counting and extrapolating data for the amount of E. coli in a sample, I was able to test my spectrophotometer.
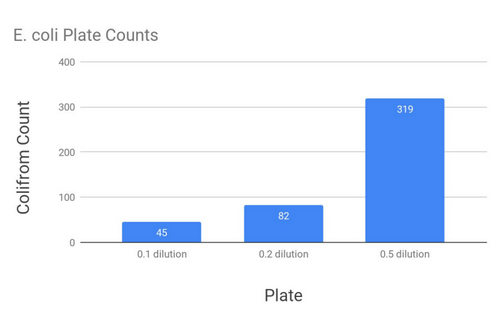
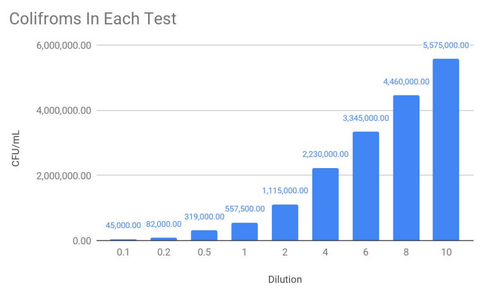
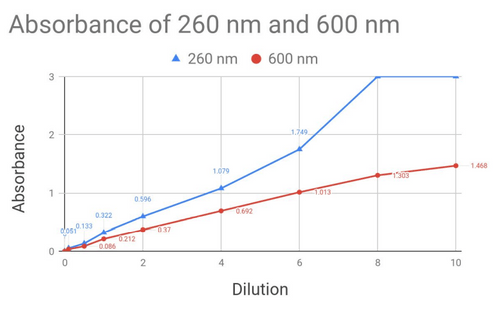
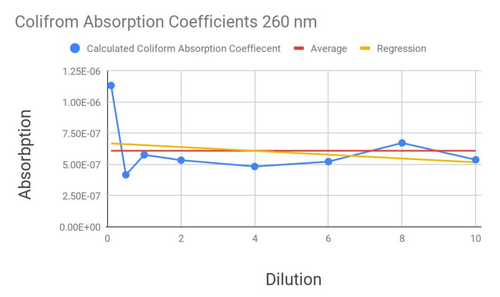
From here, I was able to calculate an absorption coefficient for E. coli (Using Beer-Lamberts Law, which tells us per coliform how much light is absorbed), which means that in a sample of unknown E. coli concentration could be tested and estimated. Testing was done with a 600nm light, using the labs spectrophotometer, to compare sensitivity.
After winning my city's science fair, I was invited to the Canada wide science fair, where I was lucky enough to win bronze in the grade 9-10 category for my work. Below is a picture of me in front of my project booth.
